All-in-one Event Management Software for
In-Person, Hybrid, and Virtual Events
All-in-one Event Management Software for
In-Person, Hybrid, and Virtual Events


Award-Winning Event App
Affordable Event Registration
Time-saving Event Management
Powerful Event Marketing
Trusted by Distinguished Innovators
Corporate Events



Academic Events

Government Events
Association Events

Conferences

Trade Shows

Festivals
& Art Shows

Make Your Attendees 10 Times Happier
Provide the best experience to your attendees with the award-winning event app! Enable them to participate and interact more actively, and build two times more connections. Up-to-date event information, personalized agenda, live polls, messages, and exciting photos are all at their fingertips.


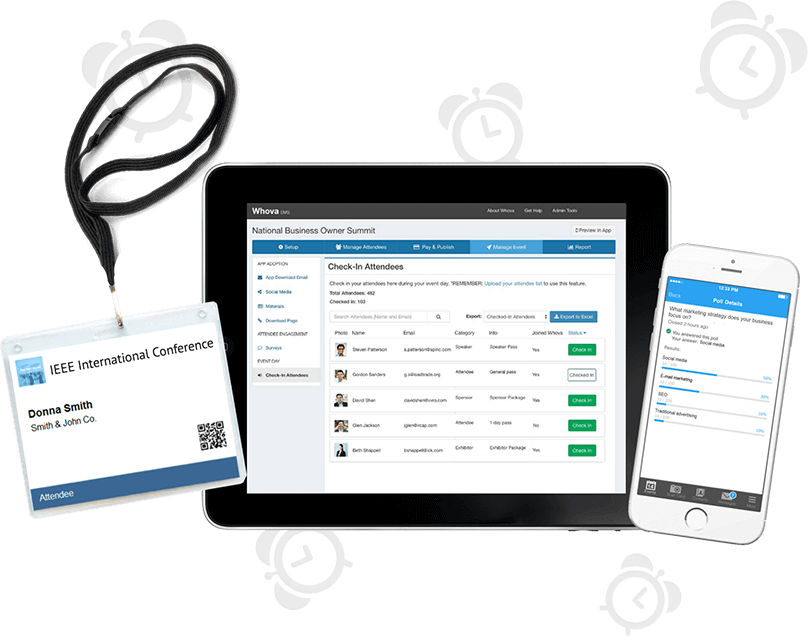
Save 60% More Time Managing Event Logistics
Complete tedious event management tasks with a few clicks and focus on more important things! Save your time with our popular event management software that includes an agenda center, speaker hub, name badge generation, check-in, announcements, and more.

Delight Sponsors and Exhibitors with Great ROI
Showcase your sponsors and exhibitors with various opportunities embedded into Whova’s event app and live slideshow. Equip them with powerful lead generation tools such as business card scanning, promotional opportunities, QR code scanning, and much more.


The Most-Loved Event App

Top-Notch Customer Support
|
Quick Responses |
|
|
Weekends and Holidays |
|
|
A Dedicated Team |

In the News
What is Unique about Whova Event Management?
Make your Event Look
Modern and Trendy
Impress your attendees, sponsors, and management team with cutting-edge technology.
Engage Attendees
Effectively
Inspire attendee loyalty by creating immersive event experiences easily and effectively
Save Time Managing
Event Logistics
Complete tedious event management tasks with a few clicks and focus on more important things!

Ready to Get Started?
It is easy to get your event set up!
Let's Work Together
We’ll find a price that fits your budget.
Let's Work Together
We’ll find a price that fits your budget.